# ppaxe
**Repository Path**: leiyis/ppaxe
## Basic Information
- **Project Name**: ppaxe
- **Description**: Text mining tool to retrieve protein-protein interactions from the scientific literature.
- **Primary Language**: Unknown
- **License**: GPL-3.0
- **Default Branch**: master
- **Homepage**: None
- **GVP Project**: No
## Statistics
- **Stars**: 0
- **Forks**: 0
- **Created**: 2020-04-15
- **Last Updated**: 2020-12-19
## Categories & Tags
**Categories**: Uncategorized
**Tags**: None
## README
[](https://doi.org/10.1093/bioinformatics/bty988)
[](https://travis-ci.org/scastlara/ppaxe)
[](https://coveralls.io/github/scastlara/ppaxe?branch=master)
[](https://badge.fury.io/py/ppaxe)
 -----
Tool to retrieve **protein-protein interactions** and calculate protein/gene symbol ocurrence in the scientific literature (PubMed & PubMedCentral). Contains two python modules (`core` and `report`), and a python script (`ppaxe`).
Available for `python 2.7` and `python 3.x`, and also as a standalone [docker image](https://hub.docker.com/r/compgenlabub/ppaxe/).
> **Visit the [PPaxe web application](https://compgen.bio.ub.edu/PPaxe) to use PPaxe on the web.**
## Citation
```
S. Castillo-Lara, J.F. Abril
PPaxe: easy extraction of protein occurrence and interactions from the scientific literature
Bioinformatics, AOP November 2018, bty988.
```
* [Link to the published paper](https://academic.oup.com/bioinformatics/advance-article-abstract/doi/10.1093/bioinformatics/bty988/5221011)
## Quick Installation
To download and use the ppaxe Docker image:
```sh
docker pull compgenlabub/ppaxe:latest
docker run -v /local/path/to/output:/ppaxe/output:rw \
compgenlabub/ppaxe -v -p ./papers.pmids -o ./output.tbl -r ./report
```
If you want to install PPaxe manually, go to the [Install ppaxe manually](#install-ppaxe-manually) section.
## Usage
```
usage: ppaxe [-h] -p PMIDS [-d DATABASE] [-o OUTPUT] [-r REPORT] [-i IP] [-v]
[-e]
Command-line tool to retrieve protein-protein interactions from the scientific
literature.
optional arguments:
-h, --help show this help message and exit
-p PMIDS, --pmids PMIDS
Text file with a list of PMids or PMCids
-d DATABASE, --database DATABASE
Download whole articles from database "PMC", or only
abstracts from "PUBMED".
-o OUTPUT, --output OUTPUT
Output file to print the retrieved interactions in
tabular format.
-r REPORT, --report REPORT
Print html report with the specified name.
-i IP, --ip IP Change the IP address of the StanfordCoreNLP server.
Default: http://localhost:9000
-v, --verbose Increase output verbosity.
-e, --exclude Exclude protein symbols not annotated in dictionary.
```
### ppaxe classes
```python
from ppaxe import core as ppcore
from ppaxe import report
# Perform query to PubMedCentral
pmids = ["28615517","28839427","28831451","28824332","28819371","28819357"]
query = ppcore.PMQuery(ids=pmids, database="PMC")
query.get_articles()
# Retrieve interactions from text
for article in query:
article.extract_interactions()
# Get the predictions
for prediction in article.predictions:
print(prediction.to_html())
# Print html report
# Will create 'report_file.html'
summary = report.ReportSummary(query)
summary.make_report("report_file")
```
### ppaxe script
```sh
# Will read PubMed ids in pmids.txt, predict the interactions
# in their fulltext from PubMedCentral, and print a tabular output
# and an html report
ppaxe -p pmids.txt -d PMC -v -o output.tbl -r report
# Or with docker image
docker run -v /local/path/to/output:/ppaxe/output:rw compgenlabub/ppaxe -v -p pmids.txt -o output.tbl -r report
```
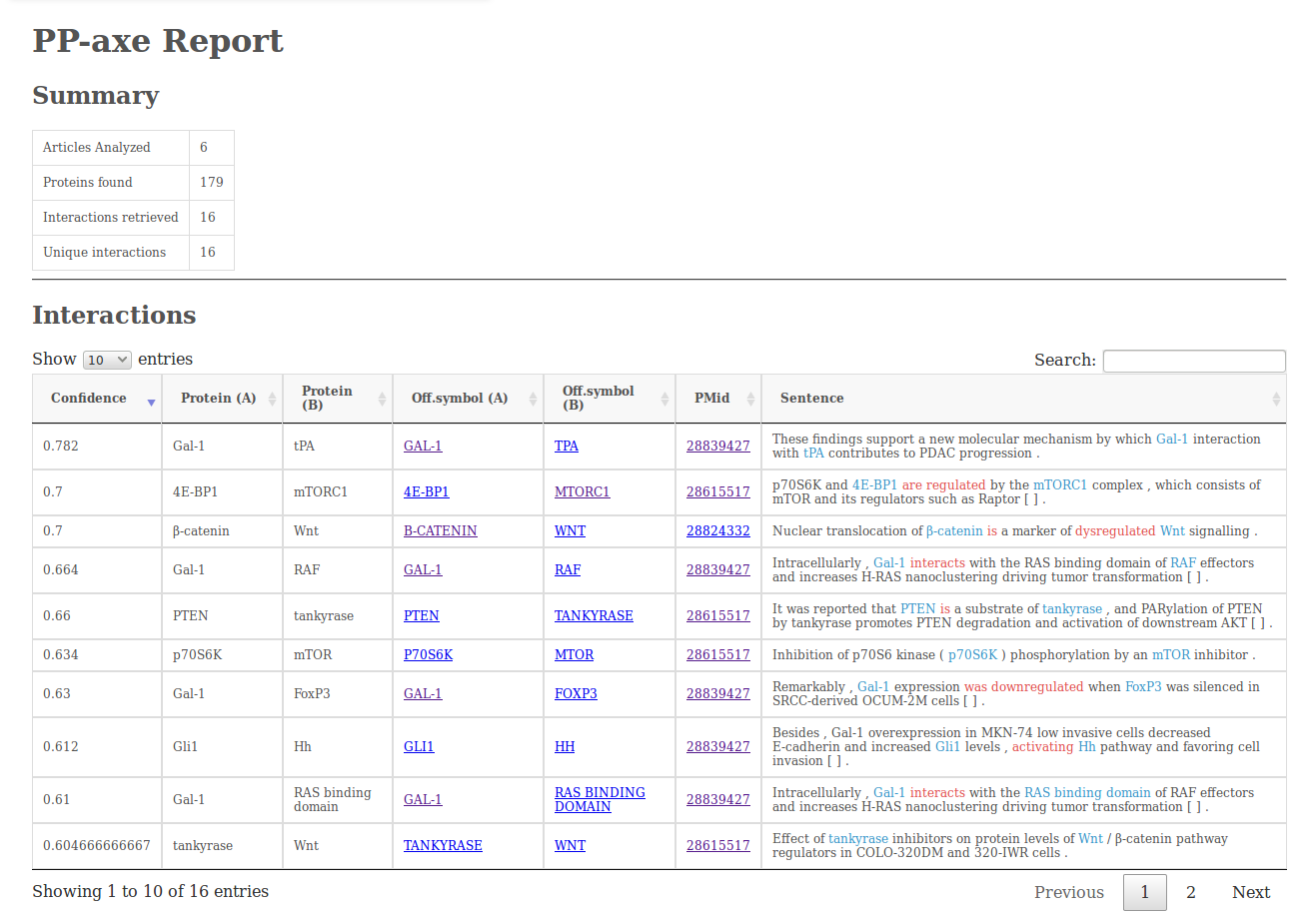
### Report
The report output (`option -r`) will contain a simple summary of the analysis, the interactions retrieved (including the sentences from which they were retrieved), a table with the protein/gene counts and a graph visualization made using [cytoscape.js](http://js.cytoscape.org/).
-----
Tool to retrieve **protein-protein interactions** and calculate protein/gene symbol ocurrence in the scientific literature (PubMed & PubMedCentral). Contains two python modules (`core` and `report`), and a python script (`ppaxe`).
Available for `python 2.7` and `python 3.x`, and also as a standalone [docker image](https://hub.docker.com/r/compgenlabub/ppaxe/).
> **Visit the [PPaxe web application](https://compgen.bio.ub.edu/PPaxe) to use PPaxe on the web.**
## Citation
```
S. Castillo-Lara, J.F. Abril
PPaxe: easy extraction of protein occurrence and interactions from the scientific literature
Bioinformatics, AOP November 2018, bty988.
```
* [Link to the published paper](https://academic.oup.com/bioinformatics/advance-article-abstract/doi/10.1093/bioinformatics/bty988/5221011)
## Quick Installation
To download and use the ppaxe Docker image:
```sh
docker pull compgenlabub/ppaxe:latest
docker run -v /local/path/to/output:/ppaxe/output:rw \
compgenlabub/ppaxe -v -p ./papers.pmids -o ./output.tbl -r ./report
```
If you want to install PPaxe manually, go to the [Install ppaxe manually](#install-ppaxe-manually) section.
## Usage
```
usage: ppaxe [-h] -p PMIDS [-d DATABASE] [-o OUTPUT] [-r REPORT] [-i IP] [-v]
[-e]
Command-line tool to retrieve protein-protein interactions from the scientific
literature.
optional arguments:
-h, --help show this help message and exit
-p PMIDS, --pmids PMIDS
Text file with a list of PMids or PMCids
-d DATABASE, --database DATABASE
Download whole articles from database "PMC", or only
abstracts from "PUBMED".
-o OUTPUT, --output OUTPUT
Output file to print the retrieved interactions in
tabular format.
-r REPORT, --report REPORT
Print html report with the specified name.
-i IP, --ip IP Change the IP address of the StanfordCoreNLP server.
Default: http://localhost:9000
-v, --verbose Increase output verbosity.
-e, --exclude Exclude protein symbols not annotated in dictionary.
```
### ppaxe classes
```python
from ppaxe import core as ppcore
from ppaxe import report
# Perform query to PubMedCentral
pmids = ["28615517","28839427","28831451","28824332","28819371","28819357"]
query = ppcore.PMQuery(ids=pmids, database="PMC")
query.get_articles()
# Retrieve interactions from text
for article in query:
article.extract_interactions()
# Get the predictions
for prediction in article.predictions:
print(prediction.to_html())
# Print html report
# Will create 'report_file.html'
summary = report.ReportSummary(query)
summary.make_report("report_file")
```
### ppaxe script
```sh
# Will read PubMed ids in pmids.txt, predict the interactions
# in their fulltext from PubMedCentral, and print a tabular output
# and an html report
ppaxe -p pmids.txt -d PMC -v -o output.tbl -r report
# Or with docker image
docker run -v /local/path/to/output:/ppaxe/output:rw compgenlabub/ppaxe -v -p pmids.txt -o output.tbl -r report
```
### Report
The report output (`option -r`) will contain a simple summary of the analysis, the interactions retrieved (including the sentences from which they were retrieved), a table with the protein/gene counts and a graph visualization made using [cytoscape.js](http://js.cytoscape.org/).
 ## Install ppaxe manually
* **Prerequisites**
```sh
xml.dom
numpy
pycorenlp
cPickle
scipy
```
You can install this package manuallly using _pip_. However, before doing so, you have to download the [Random Forest predictor](https://www.dropbox.com/s/t6qcl19g536c0zu/RF_scikit.pkl?dl=0) and place it in `ppaxe/data`.
```sh
# Clone the repository
git clone https://github.com/scastlara/ppaxe.git
# Download pickle with RF
wget https://www.dropbox.com/s/t6qcl19g536c0zu/RF_scikit.pkl?dl=0 -O ppaxe/ppaxe/data/RF_scikit.pkl
# Install
pip install ppaxe
```
* **Download StanfordCoreNLP**
In order to use the package you will need a [StanfordCoreNLP](https://stanfordnlp.github.io/CoreNLP) server setup with the [Protein/gene Tagger](https://www.dropbox.com/s/ec3a4ey7s0k6qgy/FINAL-ner-model.AImed%2BMedTag%2BBioInfer.ser.gz?dl=0).
```sh
# Download StanfordCoreNLP
wget http://nlp.stanford.edu/software/stanford-corenlp-full-2017-06-09.zip
unzip stanford-corenlp-full-2017-06-09.zip
# Download the Protein tagger
wget https://www.dropbox.com/s/ec3a4ey7s0k6qgy/FINAL-ner-model.AImed%2BMedTag%2BBioInfer.ser.gz?dl=0 -O FINAL-ner-model.AImed+MedTag+BioInfer.ser.gz
# Download English tagger models
wget http://nlp.stanford.edu/software/stanford-english-corenlp-2017-06-09-models.jar -O stanford-corenlp-full-2017-06-09/stanford-english-corenlp-2017-06-09-models.jar
# Change the location of the tagger in ppaxe/data/server.properties if necessary
# ...
# Start the StanfordCoreNLP server
cd stanford-corenlp-full-2017-06-09/
java -mx1000m -cp ./stanford-corenlp-3.8.0.jar:stanford-english-corenlp-2017-06-09-models.jar edu.stanford.nlp.pipeline.StanfordCoreNLPServer -port 9000 -serverProperties ~/ppaxe/ppaxe/data/server.properties
```
Once the server is up and running and ppaxe has been installed, you are good to go.
By default, ppaxe will assume the server is available at localhost:9000. If you want to change the address, set up the server with the appropiate port and change the address in ppaxe by assigning the new address to the variable ppaxe.ppcore.NLP:
* **Start the server**
```sh
# Change the location of the ner tagger in server.properties manually
java -mx10000m -cp ./stanford-corenlp-3.8.0.jar:stanford-english-corenlp-2017-06-09-models.jar edu.stanford.nlp.pipeline.StanfordCoreNLPServer -port your_port -serverProperties ppaxe/data/server.properties
```
* **Use the ppaxe package**
```python
from ppaxe import core as ppcore
from pycorenlp import StanfordCoreNLP
ppcore.NLP = StanfordCoreNLP(your_new_adress)
# Do whatever you want
```
## Using the Gene dictionary
By default, PPaxe uses the [HGNC](https://www.genenames.org/) dictionary of gene symbols to normalize the protein/gene symbols found in the article. The `ppaxe` command-line tool has the option `-e` that restricts all the results to only those proteins that match against the HGNC database. Users can change this file (located at `ppaxe/data/HGNC_gene_dictionary.txt`) in order to restrict their searches to only specific genes or proteins, or to normalize gene names using a different dictionary.
## Documentation
Refer to the [wiki](https://github.com/scastlara/ppaxe/wiki/Documentation) of the package.
## Running the tests
To run the tests:
```
python -m pytest -v tests
```
## Authors
* **Sergio Castillo-Lara** - at the [Computational Genomics Lab](https://compgen.bio.ub.edu)
## License
This project is licensed under the GNU GPL3 license - see the [LICENSE](LICENSE) file for details
## Install ppaxe manually
* **Prerequisites**
```sh
xml.dom
numpy
pycorenlp
cPickle
scipy
```
You can install this package manuallly using _pip_. However, before doing so, you have to download the [Random Forest predictor](https://www.dropbox.com/s/t6qcl19g536c0zu/RF_scikit.pkl?dl=0) and place it in `ppaxe/data`.
```sh
# Clone the repository
git clone https://github.com/scastlara/ppaxe.git
# Download pickle with RF
wget https://www.dropbox.com/s/t6qcl19g536c0zu/RF_scikit.pkl?dl=0 -O ppaxe/ppaxe/data/RF_scikit.pkl
# Install
pip install ppaxe
```
* **Download StanfordCoreNLP**
In order to use the package you will need a [StanfordCoreNLP](https://stanfordnlp.github.io/CoreNLP) server setup with the [Protein/gene Tagger](https://www.dropbox.com/s/ec3a4ey7s0k6qgy/FINAL-ner-model.AImed%2BMedTag%2BBioInfer.ser.gz?dl=0).
```sh
# Download StanfordCoreNLP
wget http://nlp.stanford.edu/software/stanford-corenlp-full-2017-06-09.zip
unzip stanford-corenlp-full-2017-06-09.zip
# Download the Protein tagger
wget https://www.dropbox.com/s/ec3a4ey7s0k6qgy/FINAL-ner-model.AImed%2BMedTag%2BBioInfer.ser.gz?dl=0 -O FINAL-ner-model.AImed+MedTag+BioInfer.ser.gz
# Download English tagger models
wget http://nlp.stanford.edu/software/stanford-english-corenlp-2017-06-09-models.jar -O stanford-corenlp-full-2017-06-09/stanford-english-corenlp-2017-06-09-models.jar
# Change the location of the tagger in ppaxe/data/server.properties if necessary
# ...
# Start the StanfordCoreNLP server
cd stanford-corenlp-full-2017-06-09/
java -mx1000m -cp ./stanford-corenlp-3.8.0.jar:stanford-english-corenlp-2017-06-09-models.jar edu.stanford.nlp.pipeline.StanfordCoreNLPServer -port 9000 -serverProperties ~/ppaxe/ppaxe/data/server.properties
```
Once the server is up and running and ppaxe has been installed, you are good to go.
By default, ppaxe will assume the server is available at localhost:9000. If you want to change the address, set up the server with the appropiate port and change the address in ppaxe by assigning the new address to the variable ppaxe.ppcore.NLP:
* **Start the server**
```sh
# Change the location of the ner tagger in server.properties manually
java -mx10000m -cp ./stanford-corenlp-3.8.0.jar:stanford-english-corenlp-2017-06-09-models.jar edu.stanford.nlp.pipeline.StanfordCoreNLPServer -port your_port -serverProperties ppaxe/data/server.properties
```
* **Use the ppaxe package**
```python
from ppaxe import core as ppcore
from pycorenlp import StanfordCoreNLP
ppcore.NLP = StanfordCoreNLP(your_new_adress)
# Do whatever you want
```
## Using the Gene dictionary
By default, PPaxe uses the [HGNC](https://www.genenames.org/) dictionary of gene symbols to normalize the protein/gene symbols found in the article. The `ppaxe` command-line tool has the option `-e` that restricts all the results to only those proteins that match against the HGNC database. Users can change this file (located at `ppaxe/data/HGNC_gene_dictionary.txt`) in order to restrict their searches to only specific genes or proteins, or to normalize gene names using a different dictionary.
## Documentation
Refer to the [wiki](https://github.com/scastlara/ppaxe/wiki/Documentation) of the package.
## Running the tests
To run the tests:
```
python -m pytest -v tests
```
## Authors
* **Sergio Castillo-Lara** - at the [Computational Genomics Lab](https://compgen.bio.ub.edu)
## License
This project is licensed under the GNU GPL3 license - see the [LICENSE](LICENSE) file for details -----
Tool to retrieve **protein-protein interactions** and calculate protein/gene symbol ocurrence in the scientific literature (PubMed & PubMedCentral). Contains two python modules (`core` and `report`), and a python script (`ppaxe`).
Available for `python 2.7` and `python 3.x`, and also as a standalone [docker image](https://hub.docker.com/r/compgenlabub/ppaxe/).
> **Visit the [PPaxe web application](https://compgen.bio.ub.edu/PPaxe) to use PPaxe on the web.**
## Citation
```
S. Castillo-Lara, J.F. Abril
PPaxe: easy extraction of protein occurrence and interactions from the scientific literature
Bioinformatics, AOP November 2018, bty988.
```
* [Link to the published paper](https://academic.oup.com/bioinformatics/advance-article-abstract/doi/10.1093/bioinformatics/bty988/5221011)
## Quick Installation
To download and use the ppaxe Docker image:
```sh
docker pull compgenlabub/ppaxe:latest
docker run -v /local/path/to/output:/ppaxe/output:rw \
compgenlabub/ppaxe -v -p ./papers.pmids -o ./output.tbl -r ./report
```
If you want to install PPaxe manually, go to the [Install ppaxe manually](#install-ppaxe-manually) section.
## Usage
```
usage: ppaxe [-h] -p PMIDS [-d DATABASE] [-o OUTPUT] [-r REPORT] [-i IP] [-v]
[-e]
Command-line tool to retrieve protein-protein interactions from the scientific
literature.
optional arguments:
-h, --help show this help message and exit
-p PMIDS, --pmids PMIDS
Text file with a list of PMids or PMCids
-d DATABASE, --database DATABASE
Download whole articles from database "PMC", or only
abstracts from "PUBMED".
-o OUTPUT, --output OUTPUT
Output file to print the retrieved interactions in
tabular format.
-r REPORT, --report REPORT
Print html report with the specified name.
-i IP, --ip IP Change the IP address of the StanfordCoreNLP server.
Default: http://localhost:9000
-v, --verbose Increase output verbosity.
-e, --exclude Exclude protein symbols not annotated in dictionary.
```
### ppaxe classes
```python
from ppaxe import core as ppcore
from ppaxe import report
# Perform query to PubMedCentral
pmids = ["28615517","28839427","28831451","28824332","28819371","28819357"]
query = ppcore.PMQuery(ids=pmids, database="PMC")
query.get_articles()
# Retrieve interactions from text
for article in query:
article.extract_interactions()
# Get the predictions
for prediction in article.predictions:
print(prediction.to_html())
# Print html report
# Will create 'report_file.html'
summary = report.ReportSummary(query)
summary.make_report("report_file")
```
### ppaxe script
```sh
# Will read PubMed ids in pmids.txt, predict the interactions
# in their fulltext from PubMedCentral, and print a tabular output
# and an html report
ppaxe -p pmids.txt -d PMC -v -o output.tbl -r report
# Or with docker image
docker run -v /local/path/to/output:/ppaxe/output:rw compgenlabub/ppaxe -v -p pmids.txt -o output.tbl -r report
```
### Report
The report output (`option -r`) will contain a simple summary of the analysis, the interactions retrieved (including the sentences from which they were retrieved), a table with the protein/gene counts and a graph visualization made using [cytoscape.js](http://js.cytoscape.org/).
-----
Tool to retrieve **protein-protein interactions** and calculate protein/gene symbol ocurrence in the scientific literature (PubMed & PubMedCentral). Contains two python modules (`core` and `report`), and a python script (`ppaxe`).
Available for `python 2.7` and `python 3.x`, and also as a standalone [docker image](https://hub.docker.com/r/compgenlabub/ppaxe/).
> **Visit the [PPaxe web application](https://compgen.bio.ub.edu/PPaxe) to use PPaxe on the web.**
## Citation
```
S. Castillo-Lara, J.F. Abril
PPaxe: easy extraction of protein occurrence and interactions from the scientific literature
Bioinformatics, AOP November 2018, bty988.
```
* [Link to the published paper](https://academic.oup.com/bioinformatics/advance-article-abstract/doi/10.1093/bioinformatics/bty988/5221011)
## Quick Installation
To download and use the ppaxe Docker image:
```sh
docker pull compgenlabub/ppaxe:latest
docker run -v /local/path/to/output:/ppaxe/output:rw \
compgenlabub/ppaxe -v -p ./papers.pmids -o ./output.tbl -r ./report
```
If you want to install PPaxe manually, go to the [Install ppaxe manually](#install-ppaxe-manually) section.
## Usage
```
usage: ppaxe [-h] -p PMIDS [-d DATABASE] [-o OUTPUT] [-r REPORT] [-i IP] [-v]
[-e]
Command-line tool to retrieve protein-protein interactions from the scientific
literature.
optional arguments:
-h, --help show this help message and exit
-p PMIDS, --pmids PMIDS
Text file with a list of PMids or PMCids
-d DATABASE, --database DATABASE
Download whole articles from database "PMC", or only
abstracts from "PUBMED".
-o OUTPUT, --output OUTPUT
Output file to print the retrieved interactions in
tabular format.
-r REPORT, --report REPORT
Print html report with the specified name.
-i IP, --ip IP Change the IP address of the StanfordCoreNLP server.
Default: http://localhost:9000
-v, --verbose Increase output verbosity.
-e, --exclude Exclude protein symbols not annotated in dictionary.
```
### ppaxe classes
```python
from ppaxe import core as ppcore
from ppaxe import report
# Perform query to PubMedCentral
pmids = ["28615517","28839427","28831451","28824332","28819371","28819357"]
query = ppcore.PMQuery(ids=pmids, database="PMC")
query.get_articles()
# Retrieve interactions from text
for article in query:
article.extract_interactions()
# Get the predictions
for prediction in article.predictions:
print(prediction.to_html())
# Print html report
# Will create 'report_file.html'
summary = report.ReportSummary(query)
summary.make_report("report_file")
```
### ppaxe script
```sh
# Will read PubMed ids in pmids.txt, predict the interactions
# in their fulltext from PubMedCentral, and print a tabular output
# and an html report
ppaxe -p pmids.txt -d PMC -v -o output.tbl -r report
# Or with docker image
docker run -v /local/path/to/output:/ppaxe/output:rw compgenlabub/ppaxe -v -p pmids.txt -o output.tbl -r report
```
### Report
The report output (`option -r`) will contain a simple summary of the analysis, the interactions retrieved (including the sentences from which they were retrieved), a table with the protein/gene counts and a graph visualization made using [cytoscape.js](http://js.cytoscape.org/).

 ## Install ppaxe manually
* **Prerequisites**
```sh
xml.dom
numpy
pycorenlp
cPickle
scipy
```
You can install this package manuallly using _pip_. However, before doing so, you have to download the [Random Forest predictor](https://www.dropbox.com/s/t6qcl19g536c0zu/RF_scikit.pkl?dl=0) and place it in `ppaxe/data`.
```sh
# Clone the repository
git clone https://github.com/scastlara/ppaxe.git
# Download pickle with RF
wget https://www.dropbox.com/s/t6qcl19g536c0zu/RF_scikit.pkl?dl=0 -O ppaxe/ppaxe/data/RF_scikit.pkl
# Install
pip install ppaxe
```
* **Download StanfordCoreNLP**
In order to use the package you will need a [StanfordCoreNLP](https://stanfordnlp.github.io/CoreNLP) server setup with the [Protein/gene Tagger](https://www.dropbox.com/s/ec3a4ey7s0k6qgy/FINAL-ner-model.AImed%2BMedTag%2BBioInfer.ser.gz?dl=0).
```sh
# Download StanfordCoreNLP
wget http://nlp.stanford.edu/software/stanford-corenlp-full-2017-06-09.zip
unzip stanford-corenlp-full-2017-06-09.zip
# Download the Protein tagger
wget https://www.dropbox.com/s/ec3a4ey7s0k6qgy/FINAL-ner-model.AImed%2BMedTag%2BBioInfer.ser.gz?dl=0 -O FINAL-ner-model.AImed+MedTag+BioInfer.ser.gz
# Download English tagger models
wget http://nlp.stanford.edu/software/stanford-english-corenlp-2017-06-09-models.jar -O stanford-corenlp-full-2017-06-09/stanford-english-corenlp-2017-06-09-models.jar
# Change the location of the tagger in ppaxe/data/server.properties if necessary
# ...
# Start the StanfordCoreNLP server
cd stanford-corenlp-full-2017-06-09/
java -mx1000m -cp ./stanford-corenlp-3.8.0.jar:stanford-english-corenlp-2017-06-09-models.jar edu.stanford.nlp.pipeline.StanfordCoreNLPServer -port 9000 -serverProperties ~/ppaxe/ppaxe/data/server.properties
```
Once the server is up and running and ppaxe has been installed, you are good to go.
By default, ppaxe will assume the server is available at localhost:9000. If you want to change the address, set up the server with the appropiate port and change the address in ppaxe by assigning the new address to the variable ppaxe.ppcore.NLP:
* **Start the server**
```sh
# Change the location of the ner tagger in server.properties manually
java -mx10000m -cp ./stanford-corenlp-3.8.0.jar:stanford-english-corenlp-2017-06-09-models.jar edu.stanford.nlp.pipeline.StanfordCoreNLPServer -port your_port -serverProperties ppaxe/data/server.properties
```
* **Use the ppaxe package**
```python
from ppaxe import core as ppcore
from pycorenlp import StanfordCoreNLP
ppcore.NLP = StanfordCoreNLP(your_new_adress)
# Do whatever you want
```
## Using the Gene dictionary
By default, PPaxe uses the [HGNC](https://www.genenames.org/) dictionary of gene symbols to normalize the protein/gene symbols found in the article. The `ppaxe` command-line tool has the option `-e` that restricts all the results to only those proteins that match against the HGNC database. Users can change this file (located at `ppaxe/data/HGNC_gene_dictionary.txt`) in order to restrict their searches to only specific genes or proteins, or to normalize gene names using a different dictionary.
## Documentation
Refer to the [wiki](https://github.com/scastlara/ppaxe/wiki/Documentation) of the package.
## Running the tests
To run the tests:
```
python -m pytest -v tests
```
## Authors
* **Sergio Castillo-Lara** - at the [Computational Genomics Lab](https://compgen.bio.ub.edu)
## License
This project is licensed under the GNU GPL3 license - see the [LICENSE](LICENSE) file for details
## Install ppaxe manually
* **Prerequisites**
```sh
xml.dom
numpy
pycorenlp
cPickle
scipy
```
You can install this package manuallly using _pip_. However, before doing so, you have to download the [Random Forest predictor](https://www.dropbox.com/s/t6qcl19g536c0zu/RF_scikit.pkl?dl=0) and place it in `ppaxe/data`.
```sh
# Clone the repository
git clone https://github.com/scastlara/ppaxe.git
# Download pickle with RF
wget https://www.dropbox.com/s/t6qcl19g536c0zu/RF_scikit.pkl?dl=0 -O ppaxe/ppaxe/data/RF_scikit.pkl
# Install
pip install ppaxe
```
* **Download StanfordCoreNLP**
In order to use the package you will need a [StanfordCoreNLP](https://stanfordnlp.github.io/CoreNLP) server setup with the [Protein/gene Tagger](https://www.dropbox.com/s/ec3a4ey7s0k6qgy/FINAL-ner-model.AImed%2BMedTag%2BBioInfer.ser.gz?dl=0).
```sh
# Download StanfordCoreNLP
wget http://nlp.stanford.edu/software/stanford-corenlp-full-2017-06-09.zip
unzip stanford-corenlp-full-2017-06-09.zip
# Download the Protein tagger
wget https://www.dropbox.com/s/ec3a4ey7s0k6qgy/FINAL-ner-model.AImed%2BMedTag%2BBioInfer.ser.gz?dl=0 -O FINAL-ner-model.AImed+MedTag+BioInfer.ser.gz
# Download English tagger models
wget http://nlp.stanford.edu/software/stanford-english-corenlp-2017-06-09-models.jar -O stanford-corenlp-full-2017-06-09/stanford-english-corenlp-2017-06-09-models.jar
# Change the location of the tagger in ppaxe/data/server.properties if necessary
# ...
# Start the StanfordCoreNLP server
cd stanford-corenlp-full-2017-06-09/
java -mx1000m -cp ./stanford-corenlp-3.8.0.jar:stanford-english-corenlp-2017-06-09-models.jar edu.stanford.nlp.pipeline.StanfordCoreNLPServer -port 9000 -serverProperties ~/ppaxe/ppaxe/data/server.properties
```
Once the server is up and running and ppaxe has been installed, you are good to go.
By default, ppaxe will assume the server is available at localhost:9000. If you want to change the address, set up the server with the appropiate port and change the address in ppaxe by assigning the new address to the variable ppaxe.ppcore.NLP:
* **Start the server**
```sh
# Change the location of the ner tagger in server.properties manually
java -mx10000m -cp ./stanford-corenlp-3.8.0.jar:stanford-english-corenlp-2017-06-09-models.jar edu.stanford.nlp.pipeline.StanfordCoreNLPServer -port your_port -serverProperties ppaxe/data/server.properties
```
* **Use the ppaxe package**
```python
from ppaxe import core as ppcore
from pycorenlp import StanfordCoreNLP
ppcore.NLP = StanfordCoreNLP(your_new_adress)
# Do whatever you want
```
## Using the Gene dictionary
By default, PPaxe uses the [HGNC](https://www.genenames.org/) dictionary of gene symbols to normalize the protein/gene symbols found in the article. The `ppaxe` command-line tool has the option `-e` that restricts all the results to only those proteins that match against the HGNC database. Users can change this file (located at `ppaxe/data/HGNC_gene_dictionary.txt`) in order to restrict their searches to only specific genes or proteins, or to normalize gene names using a different dictionary.
## Documentation
Refer to the [wiki](https://github.com/scastlara/ppaxe/wiki/Documentation) of the package.
## Running the tests
To run the tests:
```
python -m pytest -v tests
```
## Authors
* **Sergio Castillo-Lara** - at the [Computational Genomics Lab](https://compgen.bio.ub.edu)
## License
This project is licensed under the GNU GPL3 license - see the [LICENSE](LICENSE) file for details