代码拉取完成,页面将自动刷新



/store/wjyu/data/50Kb
library(DNAcopy)
#First, load the DNAcopy package and prepare a CNA (copy number array) object for segmentation
RT_Bg02=CNA(RTWind$Bg02esRT_.bg,RTWind$chr,RTWind$start,data.type="logratio", sampleid="RT_Bg02")
RT_ker=CNA(RTWind$keratinocytesRT_.bg,RTWind$chr,RTWind$start,data.type="logratio", sampleid="RT_ker")
#Next, segment the CNA object with the desired parameters
Seg.RT_Bg02=segment(RT_Bg02, nperm=10000, alpha = 1e-15, undo.splits ="sdundo", undo.SD=1.5, verbose=2)
Seg.RT_ker=segment(RT_ker, nperm=10000, alpha = 1e-15, undo.splits ="sdundo", undo.SD=1.5, verbose=2)
#Examine the resulting segmentation
par(ask = T,mar = c(3.1,4.1,1,1),mfrow = c(2,1))
plot(subset(Seg.RT_Bg02,chromlist = "chr1"), pch = 19, pt.cols = c("gray","gray"), xmaploc = T)
plot(subset(Seg.RT_ker,chromlist = "chr1"), pch = 19, pt.cols = c("gray","gray"), xmaploc = T)
plot(subset(Seg.RT_Bg02,chromlist = "chr2"), pch = 19, pt.cols = c("gray","gray"), xmaploc = T)
plot(subset(Seg.RT_ker,chromlist = "chr2"), pch = 19, pt.cols = c("gray","gray"), xmaploc = T)
#FIG 3
RTWind<-read.table("RT_HM.txt",header = F)
colnames(RTWind)<-c(c("chr","start","end"),list.files(path=".",pattern="*.bg"))
RTWind<-RTWind[-which(RTWind$chr=="chrY"|RTWind$chr=="chrM"|RTWind$chr=="chr*_*"),]
#Properties of RT switching domains
RT=NULL
CTCF=NULL
H3k36me3=NULL
H3k4me1=NULL
H3k4me2=NULL
H3k4me3=NULL
H3k27me3=NULL
H3k9ac=NULL
H3k27ac=NULL
RT=RTWind$Bg02esRT_.bg-RTWind$keratinocytesRT_.bg
CTCF=RTWind$Bg02esCTCF_promoter.bg-RTWind$keratinocytesCTCF_promoter.bg
H3k36me3=RTWind$Bg02esH3k36me3_promoter.bg-RTWind$keratinocytesH3k36me3_promoter.bg
H3k4me1=RTWind$Bg02esH3k4me1_promoter.bg-RTWind$keratinocytesH3k4me1_promoter.bg
H3k4me2=RTWind$Bg02esH3k4me2_promoter.bg-RTWind$keratinocytesH3k4me2_promoter.bg
H3k4me3=RTWind$Bg02esH3k4me3_promoter.bg-RTWind$keratinocytesH3k4me3_promoter.bg
H3k27me3=RTWind$Bg02esH3k27me3_promoter.bg-RTWind$keratinocytesH3k27me3_promoter.bg
H3k9ac=RTWind$Bg02esH3k9ac_promoter.bg-RTWind$keratinocytesH3k9ac_promoter.bg
H3k27ac=RTWind$Bg02esH3k27ac_promoter.bg-RTWind$keratinocytesH3k27ac_promoter.bg
HMRT<-data.frame(RTWind[,1:3],RT,CTCF,H3k36me3,H3k4me1,H3k4me2,H3k4me3,H3k9ac,H3k27me3,H3k27ac)
quantile(HMRT$RT,probs = c(0.05, 0.95),na.rm=T)
HMRT<-subset(HMRT,HMRT$RT< -3.201914 |HMRT$RT>3.011447)
cor(HMRT[,4:12],method = "spearman")
par(mfrow = c(1,1),las=2,mar=c(6,4,2,2))
cnames=c("H3k27me3","H3k9ac","H3k27ac","H3k36me3","CTCF","H3k4me3","H3k4me2","H3k4me1")
col<-matrix(c(0.11033384,0.32835090,0.36026608,0.38563788,0.44187737,0.53568363,0.5812620,0.65235889),1,8,byrow = T,dimnames=list(rnames=NULL,cnames))
barplot(col,cex.names=1,col="grey",main = "Histone Modification",ylab="Correlation (Spearman r)")
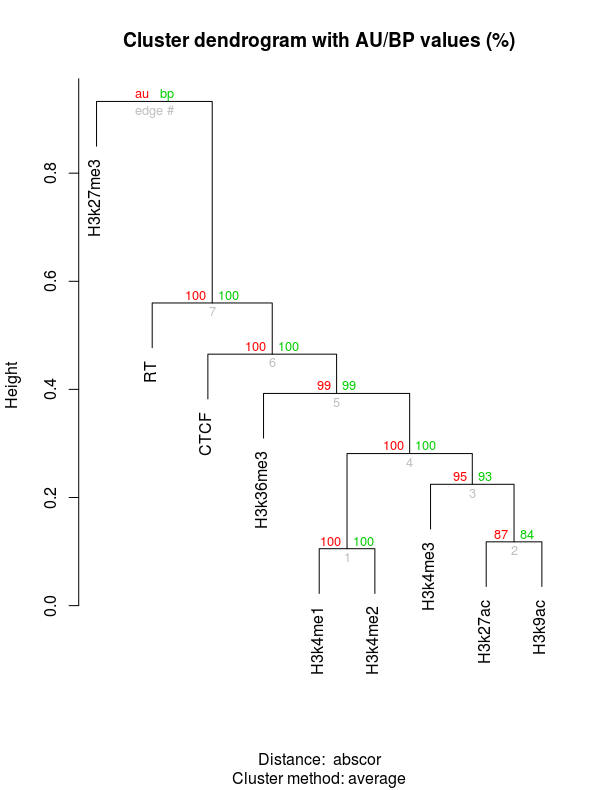
#FIG 4
cluster.bootstrap<-pvclust(RTHM[,4:12], nboot=1000, method.dist= "abscor")
plot(cluster.bootstrap)
此处可能存在不合适展示的内容,页面不予展示。您可通过相关编辑功能自查并修改。
如您确认内容无涉及 不当用语 / 纯广告导流 / 暴力 / 低俗色情 / 侵权 / 盗版 / 虚假 / 无价值内容或违法国家有关法律法规的内容,可点击提交进行申诉,我们将尽快为您处理。
登录 后才可以发表评论