代码拉取完成,页面将自动刷新
zhaosheng@nuaa.edu.cn 2021-07
依托项目
CUDA 和 NVCC 使用9.0 版本。NVCC在CUDA的bin中可以找到。
9.0的CUDA,只支持g++6.0以下的。安装g++ 4.4
切换g++版本,改变环境变量Path使用nvcc 9.0版本
Titan V: SM70 or SM_70, compute_70
DGX-1 with Volta, Tesla V100, GTX 1180 (GV104), Titan V, Quadro GV100
编译时要使用到这个参数,如果不正确会报错:cudaMemcpyToSymbol
编译
v1.5
nvcc --compiler-options -fPIE MC-GPU_v1.5b.cu -o MC-GPU_v1.5b.x -m64 -O3 -use_fast_math -DUSING_MPI -I. -I/usr/local/cuda-9.0/include -I/usr/local/cuda-9.0/samples/common/inc -I/usr/local/cuda-9.0/samples/shared/inc/ -I/usr/include/openmpi -L/usr/lib/ -lmpi -lz --ptxas-options=-v -gencode=arch=compute_70,code=sm_70 -gencode=arch=compute_70,code=sm_70
v1.3
注意:cuda-9.0的安装路径
按照官方教程会报错,要加上flag:--compiler-options -fPIE
nvcc --compiler-options -fPIE -DUSING_CUDA -DUSING_MPI MC-GPU_v1.3.cu -o MC-GPU_v1.3.x -O3 -use_fast_math -L/usr/lib/ -I. -I/usr/local/cuda-9.0/include -I/usr/local/cuda-9.0/samples/common/inc -I/usr/local/cuda-9.0/samples/shared/inc/ -I/usr/include/openmpi -L/usr/lib/ -lmpi -lz --ptxas-options=-v -gencode=arch=compute_70,code=sm_70 -gencode=arch=compute_70,code=sm_70
zubal体模的获取
zubal体模转换为penEasy(MCGPU指定的三维格式)
参考C语言代码
gcc -O3 zubal2mcgpu.c -o zubal2mcgpu.x # (Code compilation)
./zubal2mcgpu.x zubal2mcgpu_conversion_table.in voxel_man.dat # (Format conversion)
gzip voxel_man.dat.vox # (Optional compression of the phantom to save space on disk)
cd SAMPLE_SIMULATION_Zubal_phantom
../MC-GPU_v1.3.x MC-GPU_v1.3_Zubal.in | tee MC-GPU_v1.3_Zubal.out
# To visualize the output images:
gnuplot ../GNUPLOT_SCRIPTS_VISUALIZATION/gnuplot_images_MC-GPU_CT.gpl
# 两张显卡一起运行
mpirun -n 2 (后续代码...)
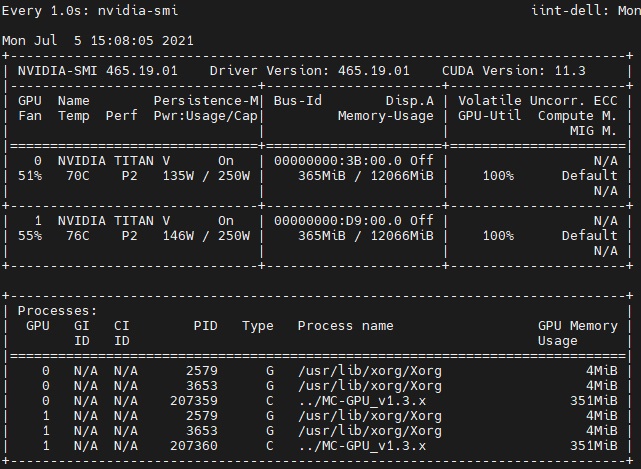
模拟速度约为 6*10^11/小时/GPU
import matplotlib.pyplot as plt
import numpy as np
import struct
from time import sleep
from tqdm import tqdm
import sys
import time
output = []
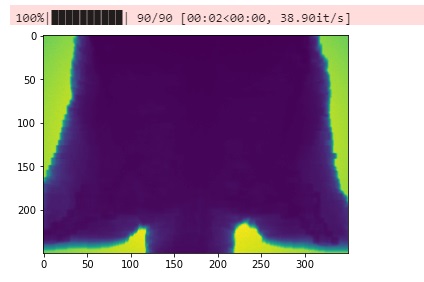
for i in tqdm(range(90)):
index = '%04d' % i
raw_file = "/home/zhaosheng/GPUMC/SAMPLE_SIMULATION_Zubal_phantom/zubal_output/zubal_test.dat_"+index+".raw"
LENGTH = 350
WIDTH = 250
HEIGHT = 5
f = open(raw_file,'rb')
data_raw = struct.unpack('f'*LENGTH*WIDTH*HEIGHT,f.read(4*LENGTH*WIDTH*HEIGHT))
data = np.asarray(data_raw).reshape(5,250,350)
pic_array = data[0,:,:]
plt.imshow(pic_array)#,cmap='gray')
output.append(pic_array[10])
assert np.array(pic_array[0]).shape == (350,)
#print(i)
#break
output = np.array(output)
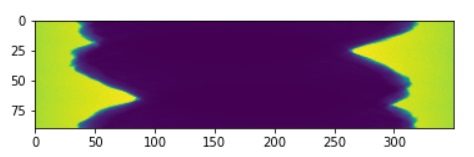
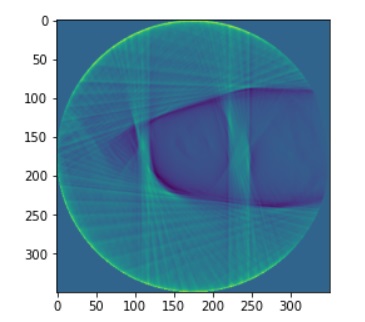
zubal.in
#
# >>>> INPUT FILE FOR MC-GPU v1.5 VICTRE-DBT >>>>>>>>>>>>>>>>>>>>>>>>>>>>>>>>>>>>>>>>>>>>>>>>>>
#
# This input file simulates a mammogram of a compressed heterogeneously dense breast phantom.
# To get simulation results fast (about 5min), the number of histories (x-ray exposure) has been set to 1% of the number of x ray estimated for a standard mammogram in this simulation conditions.
# Make sure that the folder "results" exists before starting the simulation.
#
# Main acquistion parameters:
# - Source-to-detector distance 65 cm.
# - Pixel size 85 micron (= 25.5 cm / 3000 pixels)
# - Antiscatter grid in use; no motion blur.
# - Breast phantom must be generated using C. Graff's software (hardcoded conversion from binary voxel value to material and density)
# -- It is ok to reduce the number of histories for testing the code!
# -- Number of histories computed to match the air kerma measured with the real system at the center of a PMMA phantom of equivalent thickness.
# -- Number of histotries must be re-calculated if the energy spectrum or beam aperture (field size) are changed.
#
# [Andreu Badal, 2019-08-26]
#
#[SECTION SIMULATION CONFIG v.2009-05-12]
2.0e8 # Simulating only 10% of the expected mammo exposure for testing! ORIGINAL HISTORIES: 1.7e11 # TOTAL NUMBER OF HISTORIES, OR SIMULATION TIME IN SECONDS IF VALUE < 100000
135990 # RANDOM SEED (ranecu PRNG)
0,1 # GPU NUMBER TO USE WHEN MPI IS NOT USED, OR TO BE AVOIDED IN MPI RUNS
128 # GPU THREADS PER CUDA BLOCK (multiple of 32)
5000 # SIMULATED HISTORIES PER GPU THREAD
#[SECTION SOURCE v.2016-12-02]
spectrum/W28kVp_Rh50um_Be1mm.spc # X-RAY ENERGY SPECTRUM FILE
0.1 6 50 # SOURCE POSITION: X (chest-to-nipple), Y (right-to-left), Z (caudal-to-cranial) [cm]
0.0 0.0 -1.0 # SOURCE DIRECTION COSINES: U V W
90 -90 180 # EULER ANGLES (RzRyRz) TO ROTATE RECTANGULAR BEAM FROM DEFAULT POSITION AT Y=0, NORMAL=(0,-1,0)
-1 # ==> 2/3 original angle of 11.203 # TOTAL AZIMUTHAL (WIDTH, X) AND POLAR (HEIGHT, Z) APERTURES OF THE FAN BEAM [degrees] (input negative to automatically cover the whole detector)
0.0300 # SOURCE GAUSSIAN FOCAL SPOT FWHM [cm]
0.0 # 0.18 for DBT, 0 for FFDM [Mackenzie2017] # ANGULAR BLUR DUE TO MOVEMENT ([exposure_time]*[angular_speed]) [degrees]
YES # COLLIMATE BEAM TOWARDS POSITIVE AZIMUTHAL (X) ANGLES ONLY? (ie, cone-beam center aligned with chest wall in mammography) [YES/NO]
#[SECTION IMAGE DETECTOR v.2017-06-20]
results/my__hetero_test # OUTPUT IMAGE FILE NAME
350 250 # NUMBER OF PIXELS IN THE IMAGE: Nx Nz
70.00 50.00 # IMAGE SIZE (width, height): Dx Dz [cm]
200.00 # SOURCE-TO-DETECTOR DISTANCE (detector set in front of the source, perpendicular to the initial direction)
0.0 0.0 # IMAGE OFFSET ON DETECTOR PLANE IN WIDTH AND HEIGHT DIRECTIONS (BY DEFAULT BEAM CENTERED AT IMAGE CENTER) [cm]
0.0200 # DETECTOR THICKNESS [cm]
0.004027 # ==> MFP(Se,19.0keV) # DETECTOR MATERIAL MEAN FREE PATH AT AVERAGE ENERGY [cm]
12658.0 11223.0 0.596 0.00593 # DETECTOR K-EDGE ENERGY [eV], K-FLUORESCENCE ENERGY [eV], K-FLUORESCENCE YIELD, MFP AT FLUORESCENCE ENERGY [cm]
0 # EFECTIVE DETECTOR GAIN, W_+- [eV/ehp], AND SWANK FACTOR (input 0 to report ideal energy fluence)
0.0 # ADDITIVE ELECTRONIC NOISE LEVEL (electrons/pixel)
0.10 1.9616 # ==> MFP(polystyrene,19keV) # PROTECTIVE COVER THICKNESS (detector+grid) [cm], MEAN FREE PATH AT AVERAGE ENERGY [cm]
5.0 31.0 0.0065 # ANTISCATTER GRID RATIO, FREQUENCY, STRIP THICKNESS [X:1, lp/cm, cm] (enter 0 to disable the grid)
0.00089945 1.9616 # ==> MFP(lead&polystyrene,19keV) # ANTISCATTER STRIPS AND INTERSPACE MEAN FREE PATHS AT AVERAGE ENERGY [cm]
0 # ORIENTATION 1D FOCUSED ANTISCATTER GRID LINES: 0==STRIPS PERPENDICULAR LATERAL DIRECTION (mammo style); 1==STRIPS PARALLEL LATERAL DIRECTION (DBT style)
#[SECTION TOMOGRAPHIC TRAJECTORY v.2016-12-02]
3 # ==> 1 for mammo only; ==> 25 for mammo + DBT # NUMBER OF PROJECTIONS (1 disables the tomographic mode)
60 # SOURCE-TO-ROTATION AXIS DISTANCE
45 # ANGLE BETWEEN PROJECTIONS (360/num_projections for full CT) [degrees]
0 # ANGULAR ROTATION TO FIRST PROJECTION (USEFUL FOR DBT, INPUT SOURCE DIRECTION CONSIDERED AS 0 DEGREES) [degrees]
1.0 0.0 0.0 # AXIS OF ROTATION (Vx,Vy,Vz)
0.0 # TRANSLATION ALONG ROTATION AXIS BETWEEN PROJECTIONS (HELICAL SCAN) [cm]
YES # KEEP DETECTOR FIXED AT 0 DEGREES FOR DBT? [YES/NO]
YES # SIMULATE BOTH 0 deg PROJECTION AND TOMOGRAPHIC SCAN (WITHOUT GRID) WITH 2/3 TOTAL NUM HIST IN 1st PROJ (eg, DBT+mammo)? [YES/NO]
#[SECTION DOSE DEPOSITION v.2012-12-12]
NO # TALLY MATERIAL DOSE? [YES/NO] (electrons not transported, x-ray energy locally deposited at interaction)
NO # TALLY 3D VOXEL DOSE? [YES/NO] (dose measured separately for each voxel)
mc-gpu_dose.dat # OUTPUT VOXEL DOSE FILE NAME
1 128 # VOXEL DOSE ROI: X-index min max (first voxel has index 1)
1 128 # VOXEL DOSE ROI: Y-index min max
1 243 # VOXEL DOSE ROI: Z-index min max
#[SECTION VOXELIZED GEOMETRY FILE v.2017-07-26]
../v1.3/Zubal_voxel_man.vox.gz # VOXEL GEOMETRY FILE (penEasy 2008 format; .gz accepted)
0.0 0.0 0.0 # OFFSET OF THE VOXEL GEOMETRY (DEFAULT ORIGIN AT LOWER BACK CORNER) [cm]
A 0 # NUMBER OF VOXELS: INPUT A 0 TO READ ASCII FORMAT WITH HEADER SECTION, RAW VOXELS WILL BE READ OTHERWISE
0.4 0.4 0.4 # VOXEL SIZES [cm]
0 0 0 # SIZE OF LOW RESOLUTION VOXELS THAT WILL BE DESCRIBED BY A BINARY TREE, GIVEN AS POWERS OF TWO (eg, 2 2 3 = 2^2x2^2x2^3 = 128 input voxels per low res voxel; 0 0 0 disables tree)
#[SECTION MATERIAL FILE LIST v.2009-11-30]
../v1.3/MC-GPU_material_files/air__5-120keV.mcgpu.gz # 1st MATERIAL FILE (.gz accepted)
../v1.3/MC-GPU_material_files/muscle_ICRP110__5-120keV.mcgpu.gz # 2nd MATERIAL FILE
../v1.3/MC-GPU_material_files/soft_tissue_ICRP110__5-120keV.mcgpu.gz # 3rd MATERIAL FILE
../v1.3/MC-GPU_material_files/bone_ICRP110__5-120keV.mcgpu.gz # 4th MATERIAL FILE
../v1.3/MC-GPU_material_files/cartilage_ICRP110__5-120keV.mcgpu.gz # 5th MATERIAL FILE
../v1.3/MC-GPU_material_files/adipose_ICRP110__5-120keV.mcgpu.gz # 6th MATERIAL FILE
../v1.3/MC-GPU_material_files/blood_ICRP110__5-120keV.mcgpu.gz # 7th MATERIAL FILE
../v1.3/MC-GPU_material_files/skin_ICRP110__5-120keV.mcgpu.gz # 8th MATERIAL FILE
../v1.3/MC-GPU_material_files/lung_ICRP110__5-120keV.mcgpu.gz # 9th MATERIAL FILE
../v1.3/MC-GPU_material_files/glands_others_ICRP110__5-120keV.mcgpu.gz # 10th MATERIAL FILE
../v1.3/MC-GPU_material_files/brain_ICRP110__5-120keV.mcgpu.gz # 11th MATERIAL FILE
../v1.3/MC-GPU_material_files/red_marrow_Woodard__5-120keV.mcgpu.gz # 12th MATERIAL FILE
../v1.3/MC-GPU_material_files/liver_ICRP110__5-120keV.mcgpu.gz # 13th MATERIAL FILE
../v1.3/MC-GPU_material_files/stomach_intestines_ICRP110__5-120keV.mcgpu.gz # 14th MATERIAL FILE
../v1.3/MC-GPU_material_files/water__5-120keV.mcgpu.gz # 15th MATERIAL FILE
# >>>> END INPUT FILE >>>>>>>>>>>>>>>>>>>>>>>>>>>>>>>>>>>>>>>>>>>>>>>>>>>>>>>>>
v1.5 example input
#
# >>>> INPUT FILE FOR MC-GPU v1.5 VICTRE-DBT >>>>>>>>>>>>>>>>>>>>>>>>>>>>>>>>>>>>>>>>>>>>>>>>>>
#
# This input file simulates a mammogram and 25 projections of a DBT scan (+-25deg).
# Main acquistion parameters:
# - Source-to-detector distance 65 cm.
# - Pixel size 85 micron (= 25.5 cm / 3000 pixels)
# - Antiscatter grid used only in the mammogram; motion blur used only in the DBT scan.
# - Breast phantom must be generated using C. Graff's software (hardcoded conversion from binary voxel value to material and density)
# - Number of histories matches number of x rays in a DBT projection, to reproduce the quantum noise and dose.
# -- Mammogram simulated with 2/3 the histories in the 25 projections combined (2.04e10*25*2/3 histories)
# -- It is ok to reduce the number of histories for testing the code!
# -- Number of histories computed to match the air kerma measured with the real system at the center of a PMMA phantom of equivalent thickness.
# -- Number of histotries must be re-calculated if the energy spectrum or beam aperture (field size) are changed.
#
# [Andreu Badal, 2019-08-23]
#
#[SECTION SIMULATION CONFIG v.2009-05-12]
2.04e10 # TOTAL NUMBER OF HISTORIES, OR SIMULATION TIME IN SECONDS IF VALUE < 100000
1234567890 # RANDOM SEED (ranecu PRNG)
10 # GPU NUMBER TO USE WHEN MPI IS NOT USED, OR TO BE AVOIDED IN MPI RUNS
128 # GPU THREADS PER CUDA BLOCK (multiple of 32)
5000 # SIMULATED HISTORIES PER GPU THREAD
#[SECTION SOURCE v.2016-12-02]
spectrum/W30kVp_Rh50um_Be1mm.spc # X-RAY ENERGY SPECTRUM FILE
0.00001 6.025 63.0 # SOURCE POSITION: X (chest-to-nipple), Y (right-to-left), Z (caudal-to-cranial) [cm]
0.0 0.0 -1.0 # SOURCE DIRECTION COSINES: U V W
15.0 11.203 # ==> 2/3 original angle of 11.203 # TOTAL AZIMUTHAL (WIDTH, X) AND POLAR (HEIGHT, Z) APERTURES OF THE FAN BEAM [degrees] (input negative to automatically cover the whole detector)
90.0 -90.0 180.0 # EULER ANGLES (RzRyRz) TO ROTATE RECTANGULAR BEAM FROM DEFAULT POSITION AT Y=0, NORMAL=(0,-1,0)
0.0300 # SOURCE GAUSSIAN FOCAL SPOT FWHM [cm]
0.18 # 0.18 for DBT, 0 for FFDM [Mackenzie2017] # ANGULAR BLUR DUE TO MOVEMENT ([exposure_time]*[angular_speed]) [degrees]
YES # COLLIMATE BEAM TOWARDS POSITIVE AZIMUTHAL (X) ANGLES ONLY? (ie, cone-beam center aligned with chest wall in mammography) [YES/NO]
#[SECTION IMAGE DETECTOR v.2017-06-20]
results/mcgpu_image_22183101_scattered # OUTPUT IMAGE FILE NAME
3000 1500 # NUMBER OF PIXELS IN THE IMAGE: Nx Nz
25.50 12.75 # IMAGE SIZE (width, height): Dx Dz [cm]
65.00 # SOURCE-TO-DETECTOR DISTANCE (detector set in front of the source, perpendicular to the initial direction)
0.0 0.0 # IMAGE OFFSET ON DETECTOR PLANE IN WIDTH AND HEIGHT DIRECTIONS (BY DEFAULT BEAM CENTERED AT IMAGE CENTER) [cm]
0.0200 # DETECTOR THICKNESS [cm]
0.004027 # ==> MFP(Se,19.0keV) # DETECTOR MATERIAL MEAN FREE PATH AT AVERAGE ENERGY [cm]
12658.0 11223.0 0.596 0.00593 # DETECTOR K-EDGE ENERGY [eV], K-FLUORESCENCE ENERGY [eV], K-FLUORESCENCE YIELD, MFP AT FLUORESCENCE ENERGY [cm]
50.0 0.99 # EFECTIVE DETECTOR GAIN, W_+- [eV/ehp], AND SWANK FACTOR (input 0 to report ideal energy fluence)
5200.0 # ADDITIVE ELECTRONIC NOISE LEVEL (electrons/pixel)
0.10 1.9616 # ==> MFP(polystyrene,19keV) # PROTECTIVE COVER THICKNESS (detector+grid) [cm], MEAN FREE PATH AT AVERAGE ENERGY [cm]
5.0 31.0 0.0065 # ANTISCATTER GRID RATIO, FREQUENCY, STRIP THICKNESS [X:1, lp/cm, cm] (enter 0 to disable the grid)
0.00089945 1.9616 # ==> MFP(lead&polystyrene,19keV) # ANTISCATTER STRIPS AND INTERSPACE MEAN FREE PATHS AT AVERAGE ENERGY [cm]
0 # ORIENTATION 1D FOCUSED ANTISCATTER GRID LINES: 0==STRIPS PERPENDICULAR LATERAL DIRECTION (mammo style); 1==STRIPS PARALLEL LATERAL DIRECTION (DBT style)
#[SECTION TOMOGRAPHIC TRAJECTORY v.2016-12-02]
25 # ==> 1 for mammo only; ==> 25 for mammo + DBT # NUMBER OF PROJECTIONS (1 disables the tomographic mode)
60.0 # SOURCE-TO-ROTATION AXIS DISTANCE
2.083333333333333333 # ANGLE BETWEEN PROJECTIONS (360/num_projections for full CT) [degrees]
-25.0 # ANGULAR ROTATION TO FIRST PROJECTION (USEFUL FOR DBT, INPUT SOURCE DIRECTION CONSIDERED AS 0 DEGREES) [degrees]
1.0 0.0 0.0 # AXIS OF ROTATION (Vx,Vy,Vz)
0.0 # TRANSLATION ALONG ROTATION AXIS BETWEEN PROJECTIONS (HELICAL SCAN) [cm]
YES # KEEP DETECTOR FIXED AT 0 DEGREES FOR DBT? [YES/NO]
YES # SIMULATE BOTH 0 deg PROJECTION AND TOMOGRAPHIC SCAN (WITHOUT GRID) WITH 2/3 TOTAL NUM HIST IN 1st PROJ (eg, DBT+mammo)? [YES/NO]
#[SECTION DOSE DEPOSITION v.2012-12-12]
YES # TALLY MATERIAL DOSE? [YES/NO] (electrons not transported, x-ray energy locally deposited at interaction)
NO # TALLY 3D VOXEL DOSE? [YES/NO] (dose measured separately for each voxel)
mc-gpu_dose.dat # OUTPUT VOXEL DOSE FILE NAME
1 751 # VOXEL DOSE ROI: X-index min max (first voxel has index 1)
1 1301 # VOXEL DOSE ROI: Y-index min max
250 250 # VOXEL DOSE ROI: Z-index min max
#[SECTION VOXELIZED GEOMETRY FILE v.2017-07-26]
phantom/Graff_scattered_22183101.raw.gz # VOXEL GEOMETRY FILE (penEasy 2008 format; .gz accepted)
0.0 0.0 0.0 # OFFSET OF THE VOXEL GEOMETRY (DEFAULT ORIGIN AT LOWER BACK CORNER) [cm]
1740 2415 1140 # NUMBER OF VOXELS: INPUT A 0 TO READ ASCII FORMAT WITH HEADER SECTION, RAW VOXELS WILL BE READ OTHERWISE
0.0050 0.0050 0.0050 # VOXEL SIZES [cm]
1 1 1 # SIZE OF LOW RESOLUTION VOXELS THAT WILL BE DESCRIBED BY A BINARY TREE, GIVEN AS POWERS OF TWO (eg, 2 2 3 = 2^2x2^2x2^3 = 128 input voxels per low res voxel; 0 0 0 disables tree)
#[SECTION MATERIAL FILE LIST v.2009-11-30]
material/air__5-120keV.mcgpu.gz # 1st MATERIAL FILE (.gz accepted)
material/adipose__5-120keV.mcgpu.gz # 2nd MATERIAL FILE
material/skin__5-120keV.mcgpu.gz
material/glandular__5-120keV.mcgpu.gz
material/skin__5-120keV.mcgpu.gz
material/connective_Woodard__5-120keV.mcgpu.gz
material/muscle__5-120keV.mcgpu.gz
material/muscle__5-120keV.mcgpu.gz
material/blood__5-120keV.mcgpu.gz
material/muscle__5-120keV.mcgpu.gz
material/polystyrene__5-120keV.mcgpu.gz
material/glandular__5-120keV.mcgpu.gz
material/CalciumOxalate__5-120keV.mcgpu.gz
material/W__5-120keV.mcgpu.gz
material/Se__5-120keV.mcgpu.gz # 15th MATERIAL FILE
# >>>> END INPUT FILE >>>>>>>>>>>>>>>>>>>>>>>>>>>>>>>>>>>>>>>>>>>>>>>>>>>>>>>>>
v1.3 input
# >>>> INPUT FILE FOR MC-GPU v1.3 >>>>>>>>>>>>>>>>>>>>>>>>>>>>>>>>>>>>>>>>>>>>>>>>>>
#
# Sample input file for a basic CT scan simulation:
# - 4 projections in 45 degree intervals
# - 1.0e9 histories per projection
# - 90 kVp energy spectrum
# - Full-body adult male phantom (Zubal)
#
# The Zubal male phantom can be downladed from: http://noodle.med.yale.edu/zubal/.
# The binary Zubal phantom can be converted to the MC-GPU format using the utility
# "zubal2mcgpu.c" and the conversion table "zubal2mcgpu_conversion_table.in".
#
# Voxels bounding box: 51.2 x 51.2 x 97.2 cm^3
#
# @file MC-GPU_v1.3_Zubal.in
# @author Andreu Badal (Andreu.Badal-Soler{at}fda.hhs.gov)
# @date 2012/12/12
#
#[SECTION SIMULATION CONFIG v.2009-05-12]
2.0e11 # TOTAL NUMBER OF HISTORIES, OR SIMULATION TIME IN SECONDS IF VALUE < 100000
1234567890 # RANDOM SEED (ranecu PRNG)
2 # GPU NUMBER TO USE WHEN MPI IS NOT USED, OR TO BE AVOIDED IN MPI RUNS
128 # GPU THREADS PER CUDA BLOCK (multiple of 32)
150 # SIMULATED HISTORIES PER GPU THREAD
#[SECTION SOURCE v.2011-07-12]
../90kVp_4.0mmAl.spc # X-RAY ENERGY SPECTRUM FILE
25.6 -37.0 65.0 # SOURCE POSITION: X Y Z [cm]
0.0 1.0 0.0 # SOURCE DIRECTION COSINES: U V W
-15.0 -15.0 # POLAR AND AZIMUTHAL APERTURES FOR THE FAN BEAM [degrees] (input negative to cover the whole detector)
#[SECTION IMAGE DETECTOR v.2009-12-02]
mc-gpu_image.dat # OUTPUT IMAGE FILE NAME
350 250 # NUMBER OF PIXELS IN THE IMAGE: Nx Nz
70.0 50.0 # IMAGE SIZE (width, height): Dx Dz [cm]
90.0 # SOURCE-TO-DETECTOR DISTANCE (detector set in front of the source, perpendicular to the initial direction)
#[SECTION CT SCAN TRAJECTORY v.2011-10-25]
3 # NUMBER OF PROJECTIONS (beam must be perpendicular to Z axis, set to 1 for a single projection)
45.0 # ANGLE BETWEEN PROJECTIONS [degrees] (360/num_projections for full CT)
0.0 5000.0 # ANGLES OF INTEREST (projections outside the input interval will be skipped)
60.0 # SOURCE-TO-ROTATION AXIS DISTANCE (rotation radius, axis parallel to Z)
0.0 # VERTICAL TRANSLATION BETWEEN PROJECTIONS (HELICAL SCAN)
#[SECTION DOSE DEPOSITION v.2012-12-12]
YES # TALLY MATERIAL DOSE? [YES/NO] (electrons not transported, x-ray energy locally deposited at interaction)
YES # TALLY 3D VOXEL DOSE? [YES/NO] (dose measured separately for each voxel)
mc-gpu_dose.dat # OUTPUT VOXEL DOSE FILE NAME
1 128 # VOXEL DOSE ROI: X-index min max (first voxel has index 1)
1 128 # VOXEL DOSE ROI: Y-index min max
1 243 # VOXEL DOSE ROI: Z-index min max
#[SECTION VOXELIZED GEOMETRY FILE v.2009-11-30]
Zubal_voxel_man.vox.gz # VOXEL GEOMETRY FILE (penEasy 2008 format; .gz accepted)
#[SECTION MATERIAL FILE LIST v.2009-11-30]
../MC-GPU_material_files/air__5-120keV.mcgpu.gz # 1st MATERIAL FILE (.gz accepted)
../MC-GPU_material_files/muscle_ICRP110__5-120keV.mcgpu.gz # 2nd MATERIAL FILE
../MC-GPU_material_files/soft_tissue_ICRP110__5-120keV.mcgpu.gz # 3rd MATERIAL FILE
../MC-GPU_material_files/bone_ICRP110__5-120keV.mcgpu.gz # 4th MATERIAL FILE
../MC-GPU_material_files/cartilage_ICRP110__5-120keV.mcgpu.gz # 5th MATERIAL FILE
../MC-GPU_material_files/adipose_ICRP110__5-120keV.mcgpu.gz # 6th MATERIAL FILE
../MC-GPU_material_files/blood_ICRP110__5-120keV.mcgpu.gz # 7th MATERIAL FILE
../MC-GPU_material_files/skin_ICRP110__5-120keV.mcgpu.gz # 8th MATERIAL FILE
../MC-GPU_material_files/lung_ICRP110__5-120keV.mcgpu.gz # 9th MATERIAL FILE
../MC-GPU_material_files/glands_others_ICRP110__5-120keV.mcgpu.gz # 10th MATERIAL FILE
../MC-GPU_material_files/brain_ICRP110__5-120keV.mcgpu.gz # 11th MATERIAL FILE
../MC-GPU_material_files/red_marrow_Woodard__5-120keV.mcgpu.gz # 12th MATERIAL FILE
../MC-GPU_material_files/liver_ICRP110__5-120keV.mcgpu.gz # 13th MATERIAL FILE
../MC-GPU_material_files/stomach_intestines_ICRP110__5-120keV.mcgpu.gz # 14th MATERIAL FILE
../MC-GPU_material_files/water__5-120keV.mcgpu.gz # 15th MATERIAL FILE
# >>>> END INPUT FILE >>>>>>>>>>>>>>>>>>>>>>>>>>>>>>>>>>>>>>>>>>>>>>>>>>>>>>>>>
此处可能存在不合适展示的内容,页面不予展示。您可通过相关编辑功能自查并修改。
如您确认内容无涉及 不当用语 / 纯广告导流 / 暴力 / 低俗色情 / 侵权 / 盗版 / 虚假 / 无价值内容或违法国家有关法律法规的内容,可点击提交进行申诉,我们将尽快为您处理。